On this page I want to present you different programming projects I have been working on in the last few years. Most projects will be linked to the corresponding github page. For an complete list, please take a look directly at my github profile and our labs github profile.
YAMCT (Yet Another Monte Carlo Toolbox
While there were many toolboxes developed in the last years for light simulation through Monte Carlo (MC) simulations, none of them seemed intuitive to use for me or is still based on CPU making calculations unnecessarily slow. YAMCT is an attempt to overcome those limitations by providing a user friendly toolbox. This tool will stay open source and freely accessible to everyone.
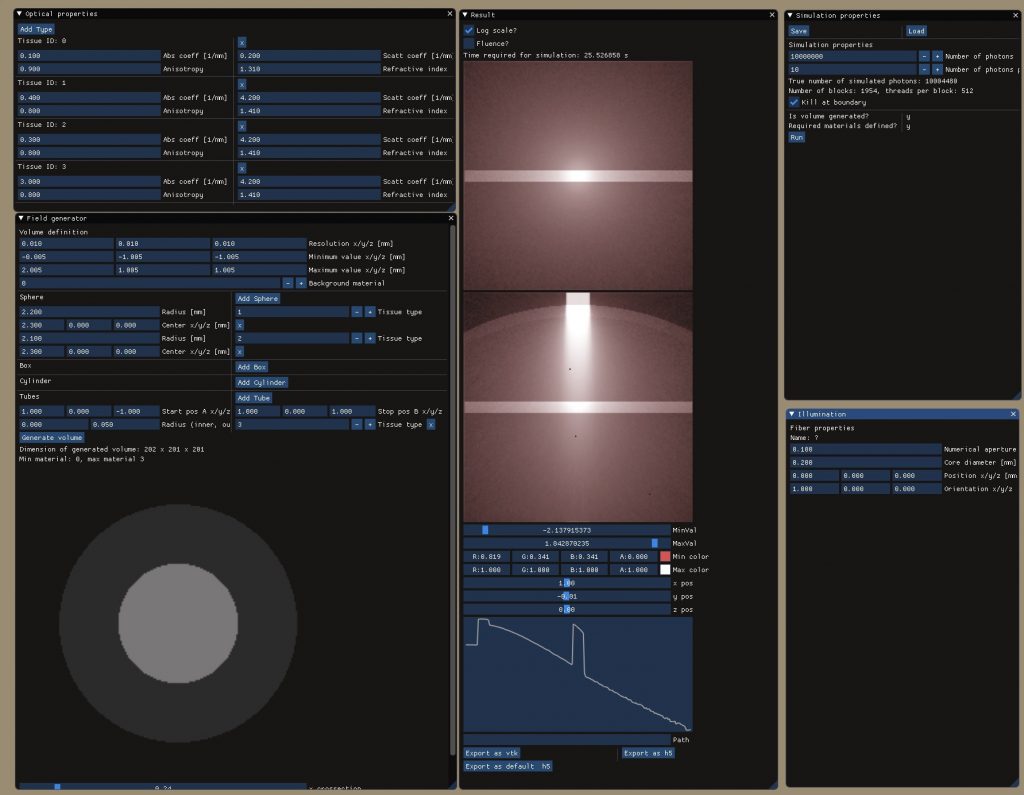
Light simulation toolbox to enable MC simulations of photons traveling through biological tissues like human skin: multilayered tissues are supported allowing the definition of arbitrarily shaped tissue blocks each material featuring its unique optical properties (refractive index, absorption coefficient, scattering coefficient, anisotropy index). Photon propagation is thereby simulated voxel by voxel for high accuracy of the calculated heat and fluence maps. Acceleration is achieved using CUDA (requires NVIDIA graphics cards) with up to 10 million photon packages being simulated in less then a minute.
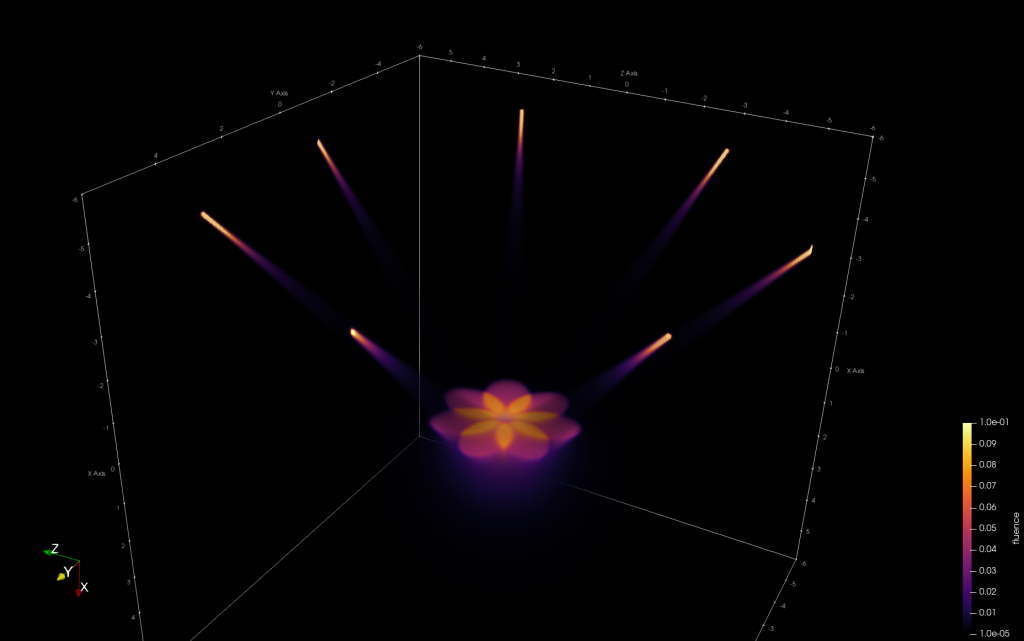
A user interface based on the amazing ImGui interface was implemented which includes interfaces to define
- different illumination profiles like multi-mode or single mode fibers at various spatial locations, orientations, and properties
- field properties like resolution and simulation volume extend
- various predefined geometrical shapes to define the tissue blocks such as tubes, spheres, boxes, etc.
- definition of optical materials including scattering coefficient, absorption coefficient etc.
- preview of results in linear / logarithmic scale as slices and plots
- file export to common data types readable through
MATLAB,Python,VTK/Paraviewetc.
Compiling is based on cmake and was tested on ArchLinux so far. Windows and Mac support is not intended but might be possible. Nevertheless, I did not try to get it running myself. Adaptation to other Linux based distributions should be straightforward. If you need help installing / compiling the toolbox for a different Linux based platform feel free to contact me. For further features, import / export options, or if you discover any bugs while using the toolbox, please contact me either here or through github.
Please note that this project is still in the development phase and while basic functionality can be guaranteed, there are still a lot of features incomplete or missing.
Links
Alternatives
opensaft
Volumetric synthetic aperture focusing technique (SAFT) for acoustic resolution optoacoustic microscopy and scanning acoustic microscopy performed in real space for arbitrary scan geometries and transducer properties. Available on github. The project was written in CUDA and C++ and mostly tested on ArchLinux so far. All dependencies should be available for all common Linux distributions though.
Features
- loading files from a standardized
*.h5file format - defining transducer geometries such as focal distance, radius of aperture, central frequency etc.
- supports both ultrasound pulse echo and optoacoustic mode
- graphical user interface based on ImGui including preview of preprocessed datasets and reconstructed datasets as slices and cropped MIPs
- export of reconstructed datasets to
*.h5and*.vtk - fully open source and free of charge (does not require MATLAB or any other commercial software)
CUDAaccelerated reconstruction procedure on GPU allows reconstruction of large datasets within seconds- Multithread processing of computationally intense calculations for on-the-fly data preview
- if requested: coherence factor weighting for improved resolution and signal-to-noise ratio
Literature
- J. Turner et al.: Improved optoacoustic microscopy through three-dimensional spatial impulse response synthetic aperture focusing technique in Optics Letters 39 (12), pp. 3390 – 3393 (2014), 10.1364/OL.39.003390
- J. Turner et al.: Universal weighted synthetic aperture focusing technique (W-SAFT) for scanning optoacoustic microscopy in Optica 4 (7), pp. 770 – 778 (2017), 10.1364/OPTICA.4.000770
- M.-L. Li et al.: Improved in vivo photoacoustic microscopy based on a virtual-detector concept in Optics Letters 31 (4), pp. 474 – 476 (2006), 10.1364/OL.31.000474
- Urs A. T. Hofmann et al.: Rapid functional optoacoustic micro-angiography in a burst mode ion Optics Letters 45 (9), pp. 2522 – 2825 (2020), 10.1364/OL.387630
MVolume
A small, object oriented interfacing class used to represent volumetric datasets in MATLAB available on github. It contains automatic definition of vectors, resolutions, and some simple export functionalities I use over and over again in my projects.
Features
- Convenient definition of datasets based on resolution and origin
- Automated frequently used dependent properties derived from
- Export of datasets into different common formats like
mat,vtkorh5
Links
CLAHE3D
Contrast limited adaptive histogram equalization to improve contrast in different regions of a volumetric datasets. Written in C++ and supporting acceleration through CUDA. The project started out as a single volume processing algorithm and now I am step by step expanding it to a broader set of functions such as
- mean filtering
- gaussian filtering
- median filtering
- thresholding
- normalization
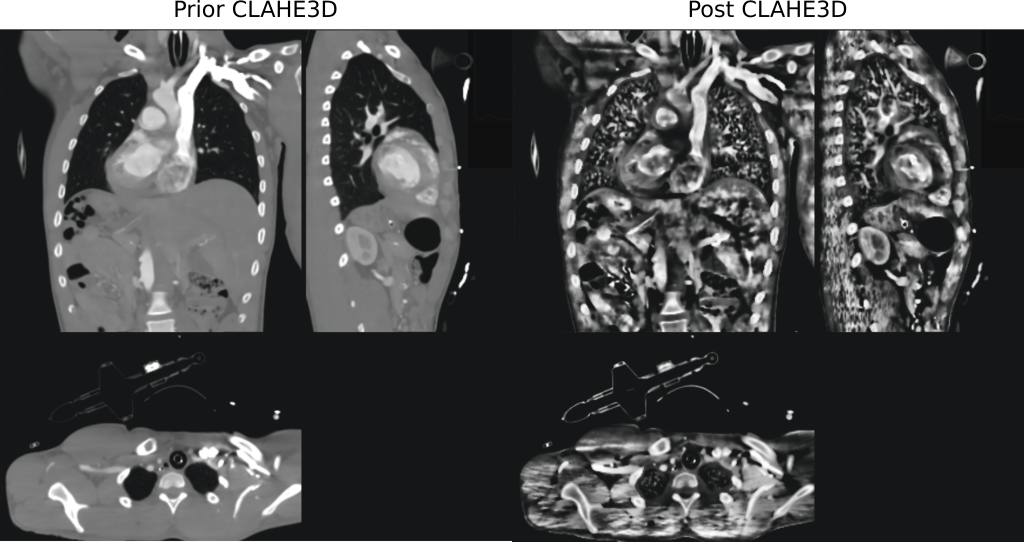
Features of the developed CLAHE3D algorithm
- overlapping regions to avoid strike artifacts at the boundaries between volumes
- basically no dependencies
- simple usage from MATLAB through
mexfunction - GPU acceleration enabled through CUDA
- including ImGUI based graphical user interface
- supports reading from standard
niiandh5files
Features of the shipped GUI
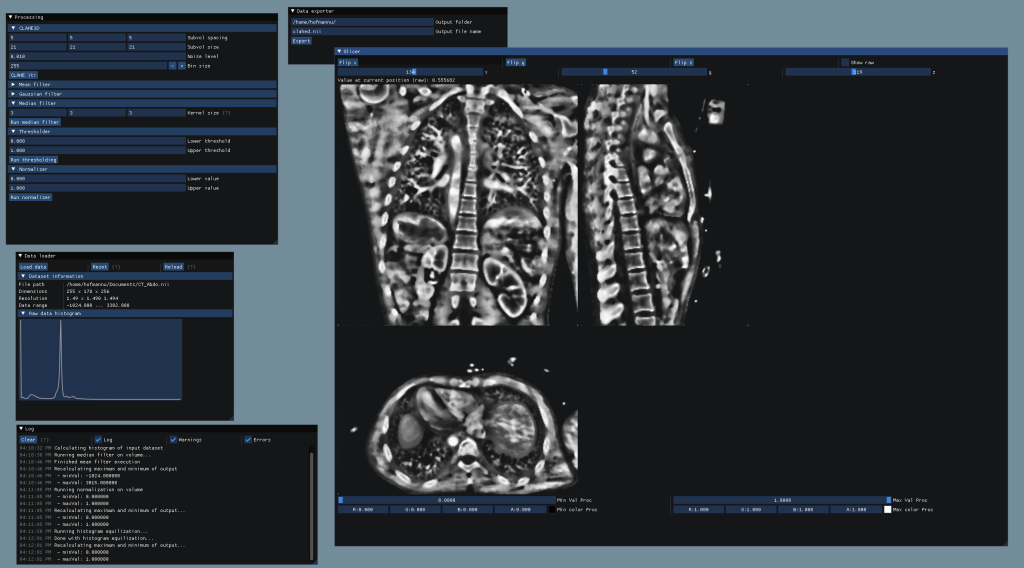
- ImGui based interface to allow simple usage of the processing algorithms
- File loading and exporting to different datatypes
- Preview of raw and processed data with included slicer
Links and Literature
- GitHub project website
- Karen Lucknavalai, Jürgen P. Schulze: Real-Time Contrast Enhancement for 3D Medical Images Using Histogram Equalization in ISVC 2020: Advances in Visual Computing pp 224-235 –> describes a layer wise processing but does not use volumetric tiling
- P Amorim et al.: 3D Adaptive Histogram Equalization Method for Medical Volumes. in VISIGRAPP (4: VISAPP), 2018
- V. Stimper et al.: Multidimensional Contrast Limited Adaptive Histogram Equalization
Hardware interfacing from MATLAB
During my PhD, most people in my lab were working with MATLAB, so I was forced to write the whole data acquisition code in that language. For each required piece of hardware I created a class and put the whole optoacoustic microscope together as a container containing those classes. Most classes can be seen as wrapper classes to interface with an existing dll or serial communication protocol.
Stages
To scan our samples or position phantoms at different locations relative to the focal point of the transducer, I used a large collection of motorized stages. Each of them requires a MATLAB interface so that we can automatically move the sample to the desired positions.

Sehr geehrter Herr Dr. Hofmann,
ich finde leider keine andere Möglichkeit als dieses Kommentarfeld, um Ihnen ein paar Fragen zu CLAHE3D zu stellen. Irgendwie gibt es Probleme beim Kompilieren auf meinem Linux-Rechner. Wie kann ich das folgende Problem lösen? Dürfte ich den Code – sofern ich ihn zum denn Laufen bewegen kann – in einem eigenen Projekt verwenden?
Viele hoffnungsvolle Grüße
[mer@cortex CLAHE3D-master]$ make main g++ -Wall -Wpedantic -ggdb -c src/histeq.cpp -o obj/histeq.o g++ -Wall -Wpedantic -ggdb -c src/interpGrid.cpp -o obj/interpGrid.o [...] g++ -Wall -Wpedantic -ggdb -c src/main.cpp -o obj/main.o src/main.cpp: In Funktion »int main()«: src/main.cpp:49:11: Fehler: »class interpGrid« hat kein Element namens »setGridSize«; meinten Sie »uint64_t interpGrid::gridSize [3]«? (aus diesem Kontext nicht zugreifbar) 49 | histGrid.setGridSize(gridSize); | ^~~~~~~~~~~ In Datei, eingebunden von src/main.cpp:11: src/interpGrid.h:9:12: Anmerkung: hier als »private« deklariert 9 | uint64_t gridSize[3]; // size of subvolume grid | ^~~~~~~~ make: *** [makefile:13: obj/main.o] Fehler 1 [mer@cortex CLAHE3D-master]$Natuerlich koennen Sie den Code verwenden. Ich habe den Fehler entsprechend korrigiert. Bitte benutzen Sie die neueste Version auf GitHub.
Hallo Herr Dr. Hofmann,
nach längerer Pause melde ich mich mal wieder. Mir aufgefallen, dass die abgespeicherten NIFTI-Dateien nicht lesbar sind:
[mer@cortex build (master)]$ fslinfo test_clahe.nii
Error: file does not appear to be a valid NIFTI or ANALYZE image
[mer@cortex build (master)]$
Viele Grüße